Strategic Partners for the Evaluation of Predictive Signatures of Prostate Cancer (SPECS)
Executive Summary
The Goal of the UCI SPECS program in prostate cancer is to develop cell-type specific multigene signatures for the prognosis of prostate cancer at the time of diagnosis. A unique feature of our approach is to identify gene expression as measured by Affymetrix GeneChips for particular cell types that make up the complex tissue of prostate cancer (tumor epithelial cells, epithelial cells of benign prostate hyperplasia, epithelial cell of dilated cystic glands, and stroma cells). We have recruited over 1500 patients from seven different sites in Southern California provided by our SPECS consortium partners at UCI, UCSD, and SKCC, and from our partners at the SPORE of North Western University to obtain tissues for this analysis and developed specialized bioinformatics approaches to extract the predictive information. Frozen tissue with clinical annotation was used to develop a signature predictive of the outcome of prostate cancer at the time of diagnosis. Observed gene expression is expressed as a linear model of the four contributing cell types.
![]()
The gene expression of each cell type is then extracted by the use of multiple linear regression analysis. Differential expression of tumor cells between relapsed disease as judged by chemical failure and nonrelapsed cases provides a signature of "aggressive" disease. Application of these signatures to new "test" cases accurately provides an estimate risk of relapse at the time of diagnosis. 148 GeneChips have been used in the training phase, and 108 GeneChips of separate cases served as the test set. Prognostic gene signatures (i) may guide treatment choice, (ii) determine who may best profit from radical treatment such as prostatectomy, and (iii) who may profit from postsurgery adjuvant therapy. Independent experimental validation of these results is underway.
To develop a clinically and commercially useful test from these results, a virtual entity, Proveri Inc. (www.provericorp.com) has been developed which has obtained a world-wide exclusive license from the Regents of the University of California, the SPECS grant holder. For translation, a sublicense has been provided by Althea Technologies Inc. of San Diego (www.altheatech.com) which specializes in the use of multiplex PCR and are developing a kit for the prediction of outcome based on the use of preserved hospital tissues of patients, a resource that exists for virtually all patients that receive a diagnosis of prostate cancer (for SPECS and Althea see http://altheadx.com/prostate-cancer-prognostic-assay.php). The test is to be applied prior to therapy as a guide to who needs radical and potentially dangerous procedures and of those who have surgery, who may profit from immediate surgery adjuvant therapy, an option rarely offered today.
Diagnosis. Recently we have developed a new "diagnostic" gene signature that detects the presence of prostate cancer solely from the gene expression changes in the "microenvironment" (tumor adjacent stroma) of tumors, that is, detection of tumor in the absence of tumor cells. This test is badly needed to clarify the examination of prostate biopsies. Over 1 million biopsies are performed in the U.S. annually and 10-15% are found to be "nondiagnostic" owing to too little tumor for a definitive interpretation or due to the presence of "atypical" glands only or other caveats. Thus a test that exploits the hundreds of abnormal gene expression changes that we observed in the tumor microenvironment will help clarify these cases. The use of tumor microenvironment is a novel approach to diagnosis. We used rigorous methods such as the development of an entirely independent set of 108 pangenomic Affymetrix GeneChips arrays as a "test" set of cases to validate our test which yields 85% accuracy. For translation, Proveri Inc. has applied for an SBIR with Vala Sciences Inc. (www.valasciences.com) of San Diego, who specialize in rapid tissue scanning by immunofluorescence for the development of an antibody panel targeting gene products of our gene signature. Proveri and Vala will co-develop the antibody assay including antibody-based kit production. In addition Althea Technologies is actively exploring the potential of an RNA-based multiplex PCR diagnostic assay.
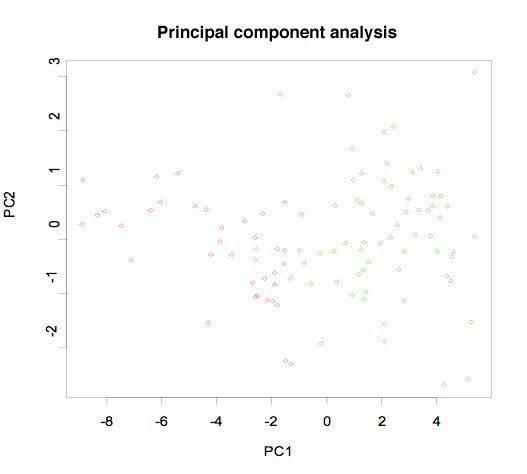
The graph illustrates an application of a 42 diagnosis gene "classifier" profile to separate prostate cancer cases from nontumor tissues of our "test" set of 91 cases analyzed on 108 Affymetrix U133 plus 2 GeneChips using here principal component analysis. The 42 gene classifier is 94% accurate.
Insight. The genes of the prognostic and diagnostic signatures may provide important clues to the function of prostate cancer and may be useful targets of therapy.
Resources. To further validate our results we work with our SPECS consortium partners at The Burnham Institute to make tissue microarrays now containing 272 cases. By using antibodies to the genes of largest expression changes, we can confirm on the independent cases of the TMA and at the protein level that our signatures are correct. The arrays are unique and contain "cores" from each case of pure cell types, have been vetted by two pathologists, and are clinically annotated for outcome, and many related conditions of prostate cancer in our clinical database. These arrays contain cases of normal prostate tissue for control purposes that have been provided by our SPECS consortium partners at The Sun Health Research Institute who carry out a rapid autopsy program. Normal tissue controls are rare in prostate cancer research and these are an important addition to our TMAs. All cases have extensive clinical annotation maintained in the SPECS database as organized by Manuel Sutton and Anne Sawyers. Another 150 clinically annotated cases are currently being applied to the array. The interim TMAs have been used in two publications.
Many of the 1500 consented cases have banked frozen plasma and post DRE-urine. Over 500 frozen research biopsies have been banked. Currently frozen research biopsies are being received from the University of Surrey, U.K. Materials banked since the inception of the study, Sept. 2005, to date are useful for prospective validation of our results as well as others.
310 Affymetrix GeneChips have been analyzed for samples with known cell-type compositions (147 U133A, deposited with GEO (Accession GSE8218); 108 U133 plus 2; 54 Exon). 77 Illumina SNP arrays have been completed.
The SPECS database contains extensive clinical annotation for up to 19 years for over 1500 cases. The database is the sole resource for Ph.D. dissertation studies of J. Major, M.S., who is examining gene correlates of diabetes and dietary factors in prostate cancer.
Supplementary Funding. G. Saxe of UCSD has provided a three year subcontract of $100,000 for the study of gene expression correlates of diabetes and obesity. The UCI Cancer Center has provided $35,000 per year for the development of a CaBIG compatible version of the SPECS clinical database. The United Kingdom has provided a three year "SETsquared" grant of $40,000 per year for the analysis of frozen biopsies of U.K. National Health prostate cancer patients. Two Ph.D. students and two postdoctoral scholars are full-time investigators on SPECS projects but largely supported by separate funding of their mentors. Anne Simoneau, a surgical urologist at UCI, has provided fresh frozen research biopsies of normal subjects without charge. Income from the Althea License covers the Proveri license maintenance costs and will produce loyalty income upon product development milestone completion.
Ph.D. Students and Fellows. Jacqueline Major, UCSD, analysis of gene study of gene expression correlates of diabetes and obesity. Rebecca Pio, UCI, analysis of DNA regulation by transcription factors identified in generated resources. Four postdoctoral scholars, Yipeng Wang, Tolga Turan, Shilpi Arora, and Farah Raymatpanah, are supervised by SPECS site PIs and focused on functional aspects arising from observations of gene expression of the SPECS study.
Patents Pending. A patent application covering the methods and genes licensed by Proveri Inc. has been applied for by the Regents of the University of California (USPTO #20060292572; iEdison EIR no. 2560101-03-0010). A data sharing agreement is enforced among all participating institutes of the UCI SPECS consortium. (http://tinyurl.com/25zr3ot)
Publications and Presentations. Six publications (1-6) and nine published abstracts of presentations have been completed (7-15).
Pending Funding Activity. Four SBIR applications and two DOD interactive IDEA applications are pending.
